How well can today’s leading GenAI models classify real-world biodiversity data—without bespoke code or traditional machine learning pipelines? In this study, we benchmarked a range of large language models on the task of predicting penguin species from tabular ecological measurements, including both numerical and categorical variables. Using a set of 100 labelled training samples and 30 test cases, we assessed each model’s ability to assign the correct species based on the available attributes. Grok 3, DeepSeek-V3, and Qwen2.5-Max all stood out with perfect prediction accuracy, while Claude 4 Opus, Mistral, Gemini 2.5 Pro, and GPT-4.1 (and GPT-4o) also achieved high accuracy, with only a small number of misclassifications. In contrast, Copilot was unable to complete the task and lagged significantly behind the other models. This experiment demonstrates the remarkable progress in prompt-based, code-free data analysis using large language models, and points to promising new directions for rapid and reproducible scientific workflows.
Input files
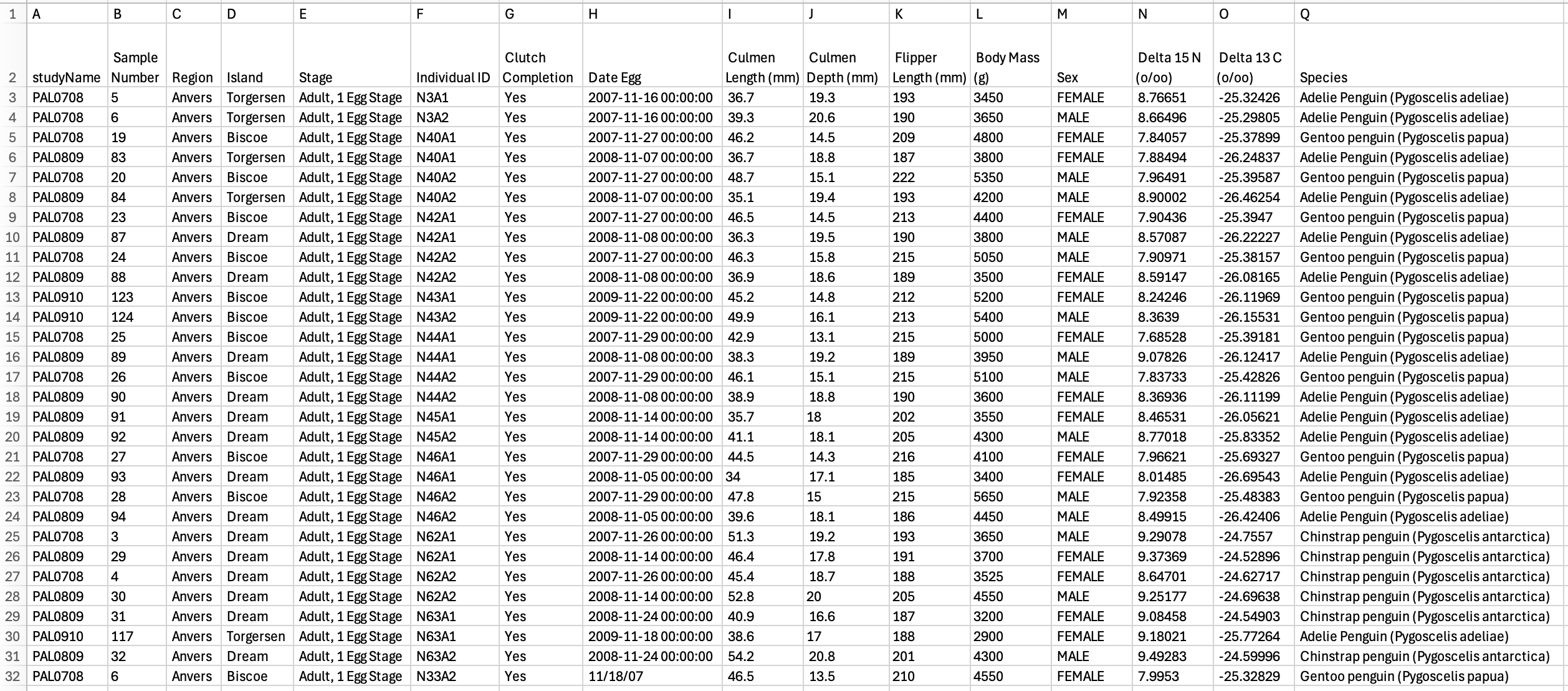
The data for this experiment were sourced from the public Kaggle project “Penguin Clustering (EDA+PCA+Kmeans)”. The dataset contains comprehensive ecological measurements for individual penguins, including species labels and a range of biometric and categorical attributes (such as culmen length, flipper length, body mass, sex, and island). For the purposes of this benchmark, we extracted 100 labelled samples as the training set and selected 30 additional cases as a held-out test set, preserving the original feature structure.
Prompt
The challenge for each model was to accurately classify the species of penguins in the test set using only the patterns and relationships found within the 100 labelled training examples. Models received no external hints or prior knowledge—just the raw tabular data with features including physical measurements, island, and sex. For each of the 30 unlabelled test cases, the model was asked to append its predicted species as a new column, producing a completed test table identical in format to the training set. The evaluation focused strictly on the model’s ability to “learn” from these examples in context and generalise to new cases, without explanations, commentary, or changes to the table structure—just a straightforward, row-by-row prediction based on the data supplied.
You are provided with two Excel spreadsheets, both derived from the Palmer Penguins dataset:
penguins_train.xlsx: Contains 100 labelled examples, with the following columns:
Culmen Length (mm), Culmen Depth (mm), Flipper Length (mm), Body Mass (g), Island, Sex, and the target column Species.penguins_test.xlsx: Contains 30 unlabelled examples in the exact same format, but the Species column is missing.
Task:
Using only the information in the 100 labelled examples, learn how the physical measurements (Culmen Length (mm), Culmen Depth (mm), Flipper Length (mm), Body Mass (g)), the island, and the sex relate to the species of penguin.
There are three possible species to assign:
- Adelie Penguin (Pygoscelis adeliae)
- Gentoo penguin (Pygoscelis papua)
- Chinstrap penguin (Pygoscelis antarctica)
Your task is to predict the correct Species for each of the 30 test examples. For each test row, insert your prediction as a new column, in the same format as the training data, so that the output table contains all the original columns plus the predicted Species column at the end.
Instructions:
- Base your predictions solely on the patterns, boundaries, and relationships you observe in the training data.
- Do not use any outside knowledge or make assumptions beyond what is present in the 100 training rows.
- Output only the completed table, with the predicted Species column appended as the final column for each test row.
- Preserve the original order and formatting of the test table.
- Do not provide explanations or commentary—just return the completed table.
Output
The table below summarises the prediction accuracy of each tested GenAI model on the 30-row penguin species classification benchmark, clearly highlighting which models achieved the highest scores and which fell short.
| Model | Correct Predictions (out of 30) | Accuracy (%) | Notes |
|---|---|---|---|
| Grok 3 | 30 | 100% | Perfect accuracy, all predictions correct |
| DeepSeek-V3 | 30 | 100% | Perfect accuracy, all predictions correct |
| Qwen2.5-Max | 30 | 100% | Perfect accuracy, all predictions correct |
| Claude 4 Opus | 26 | 86.7% | 4 misclassifications |
| GPT-4.1 (and GPT-4o) | 26 | 86.7% | 4 misclassifications |
| Mistral | 25 | 83.3% | 5 misclassifications |
| Gemini 2.5 Pro | 24 | 80% | 6 misclassifications |
| Copilot | 0 | 0% | Unable to complete the task |
Grok 3, DeepSeek-V3, and Qwen2.5-Max
The three top-performing models—Grok 3, DeepSeek-V3, and Qwen2.5-Max—each achieved perfect (100%) accuracy in the penguin species classification task. Since its initial release, Grok 3 has undergone several upgrades; previously it could not generate downloadable files, but this feature is now supported, making it the most straightforward and accurate solution in our evaluation, with results that are immediately exportable. A particularly useful feature of the Grok interface is the ability to preview the generated output file directly within the platform. By clicking “Show inline,” users can expand and view the content of the completed CSV table in their browser, without needing to download and open the file separately.

DeepSeek-V3, Qwen2.5-Max and Mistral Le Chat
By contrast, DeepSeek-V3 and Qwen2.5-Max are open-source models. Although exporting their results to Excel or CSV requires an extra step, they also delivered 100% accuracy, and their transparency and free availability are significant advantages for research and reproducibility.
Among the models with a small number of misclassifications, Claude 4 Opus and Gemini 2.5 Pro were unable to produce downloadable output files, requiring manual copying of the table from the interface. In contrast, GPT-4.1 (and GPT-4o) as well as Mistral did generate downloadable files, streamlining the workflow. Notably, Mistral stands out as an open-source model, offering both transparency and ease of access for further research or integration.
Copilot
In Copilot’s case, the workflow was hampered by technical limitations: it did not allow both the train and test files to be uploaded at the same time, as each new upload replaced the previous one. Even after both files had been re-uploaded and the model was prompted several times, Copilot repeatedly replied that it was “working on it” and would provide the completed table as soon as possible. However, even after more than an hour, no output was produced and the model did not return any predictions for the test set.
Recommendations
This experiment shows that several current GenAI models can accurately classify species from tabular ecological data using prompt-based instructions and a modest training set. Nevertheless, workflow differences remain important: some models—such as Grok 3, GPT-4.1 (and 4o), and Mistral—offer convenient downloadable output formats, streamlining integration with other analysis tools. Others, including DeepSeek-V3, Qwen2.5-Max, Claude 4 Opus, and Gemini 2.5 Pro, require manual copying of results from the interface, which may slow down the process or introduce errors. Copilot, meanwhile, was unable to process the task under these conditions. For applied research, we recommend testing the full workflow in advance and verifying predictions against ground truth data, as both technical features and predictive performance may vary between models.
The authors used Grok 3 [xAI (2025) Grok 3 (accessed on 30 May 2025), Large language model (LLM), available at: https://x.ai/grok] to generate the output.






