Working with biomedical corpora often requires programming skills, specialised formats, and time-consuming preprocessing. But what if you could transform a complex annotated dataset—like the NCBI Disease Corpus—into a structured, analysis-ready CSV using nothing more than a single, well-designed prompt? In this post, we demonstrate how a no-code, GenAI-powered approach can extract titles, abstracts, and disease mentions from the raw corpus file and reorganise them into a clean, tabular format—without writing a single line of code. In this post, we demonstrate how GPT-4o was able to perform this task with no code involved, successfully converting unstructured annotation files into a clean, spreadsheet-ready format. The example highlights the practical capabilities of large language models when applied to real-world data structuring challenges.
Input file
The dataset used in this transformation task is the NCBI Disease Corpus, a manually annotated biomedical corpus developed by the National Center for Biotechnology Information (NCBI). While the full corpus contains 793 PubMed abstracts and over 6,800 disease mentions, for this task we worked specifically with the training set, which includes 592 abstracts. Each disease mention is labelled with precise character offsets and linked to standard biomedical vocabularies such as MeSH or OMIM.
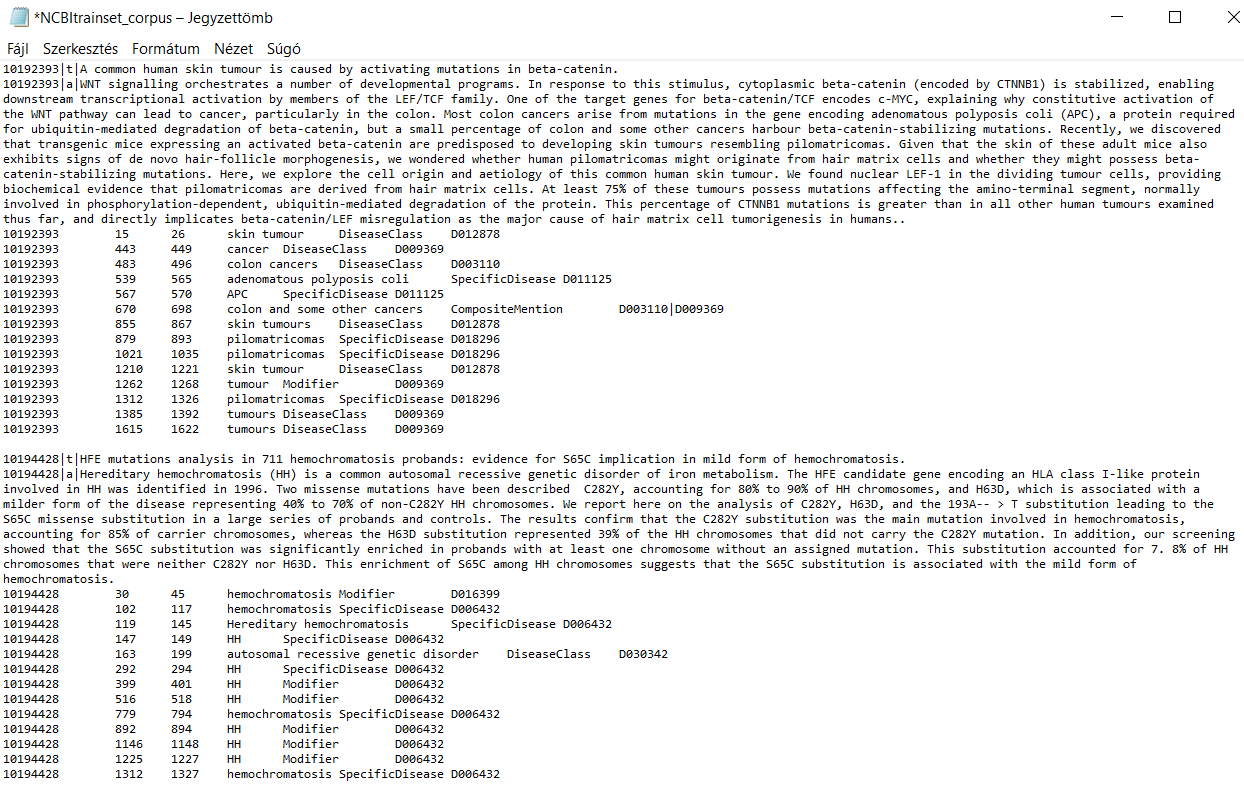
The file used (NCBItrainset_corpus.txt) follows a structured plain-text format, where each block consists of:
- a title line, marked with
|t| - an abstract line, marked with
|a| - a series of annotation lines, each with six tab-separated fields:
PMID,start offset,end offset,mention text,entity type, andMeSH ID
While this format is compact and machine-readable, it is not immediately suitable for spreadsheet-based analysis. We aimed to convert it into a clean, structured CSV with one row per article and all disease mentions as separate columns. Below is a sample showing how the data is structured in its original format.
Prompt
Although the original file contains a wide range of metadata—including character offsets, entity types, and MeSH identifiers—our goal in this task was not to extract every available detail. Instead, we focused on retrieving only the information necessary to create a structured and readable summary of each abstract.
The prompt was designed to return:
- the title of each entry,
- the corresponding abstract, and
- all annotated disease mentions, listed in separate columns (e.g. Disease1, Disease2, etc.).
We asked the model to process every annotation line in the file, regardless of entity type—including those labelled as Modifier—to ensure that no mention was omitted. The output was expected in a clean, comma-separated format suitable for direct export to Excel or other tabular environments.
You have access to a file I have uploaded named "NCBItrainset_corpus.txt".
This file contains annotated biomedical data from the NCBI Disease Corpus. Your task is to transform this file into a structured format suitable for export to Excel.
Each abstract block consists of:
- One title line marked with "|t|"
- One abstract line marked with "|a|"
- One or more annotation lines, each tab-delimited with the following fields: PMID, start offset, end offset, mention text, entity type, MeSH ID
Please do the following:
- For each unique PMID, extract:
- The title from the "|t|" line
- The abstract from the "|a|" line
- All disease mentions from the annotation lines. You must include every single annotated mention, regardless of its entity type. That includes mentions with the entity type "Modifier". Do not filter or exclude any lines.
- Build a table with the following columns:
- Column A: An ID starting from 1
- Column B: The extracted title
- Column C: The abstract
- Columns D and onwards: One column for each disease mention, in the order they appear (e.g. Disease1, Disease2, Disease3, ...)
The result must be formatted as comma-separated values (CSV-compatible), one row per abstract. Do not include any commentary, explanation or additional text before or after the output.
Important: Do not ignore or exclude any annotated lines. All lines must be processed, even if the entity type is "Modifier".
Output
The output generated by GPT-4o was produced in just a few seconds and met all the requirements specified in the prompt. The model successfully extracted the titles, abstracts, and all disease mentions—regardless of entity type—and arranged them into a structured, CSV-compatible format.
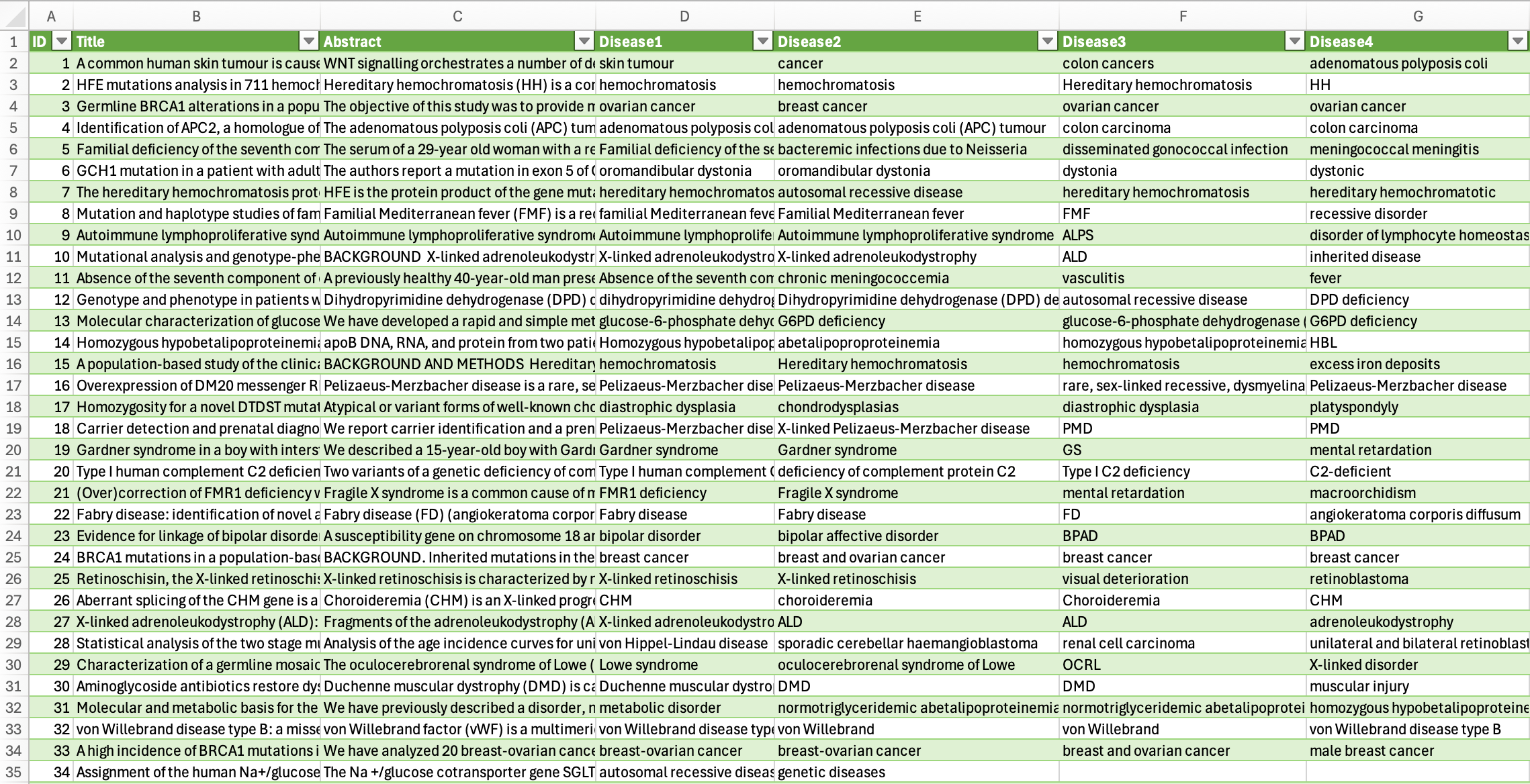
To verify the quality of the transformation, we conducted a manual validation on a random sample of 10 abstracts. In each case, the extracted data matched the original annotations perfectly, with no omissions or formatting errors. This gave us high confidence in the result's reliability. The resulting CSV file is now ready for further use in data analysis, visualisation, or integration into other workflows—no post-processing or corrections were necessary.
Recommendations
This case shows that with a well-crafted prompt, even complex biomedical corpora like the NCBI Disease Corpus can be reliably restructured using a no-code approach. GPT-4o produced clean, accurate output in seconds, requiring no further corrections. We recommend prompt-based methods for similar data preparation tasks—especially when transparency, speed, and minimal technical setup are priorities.
The authors used GPT-4o [OpenAI (2025), GPT-4o (accessed on 27 May 2025), Large language model (LLM), available at: https://openai.com] to generate the output.






