Recent advancements in multimodal language models have opened new avenues for analysing scientific image data using natural language instructions. In this post, we explore the capabilities of OpenAI’s o4-mini-high model for performing cell segmentation tasks on microscopy images through prompt-based interaction. Rather than relying on traditional computer vision techniques or fully supervised deep learning models, we investigate whether a lightweight, general-purpose model can generate meaningful binary masks solely from descriptive prompts. We compare the prompt-driven results with classical OpenCV-based segmentation and assess their usefulness for biomedical imaging workflows.
Input files
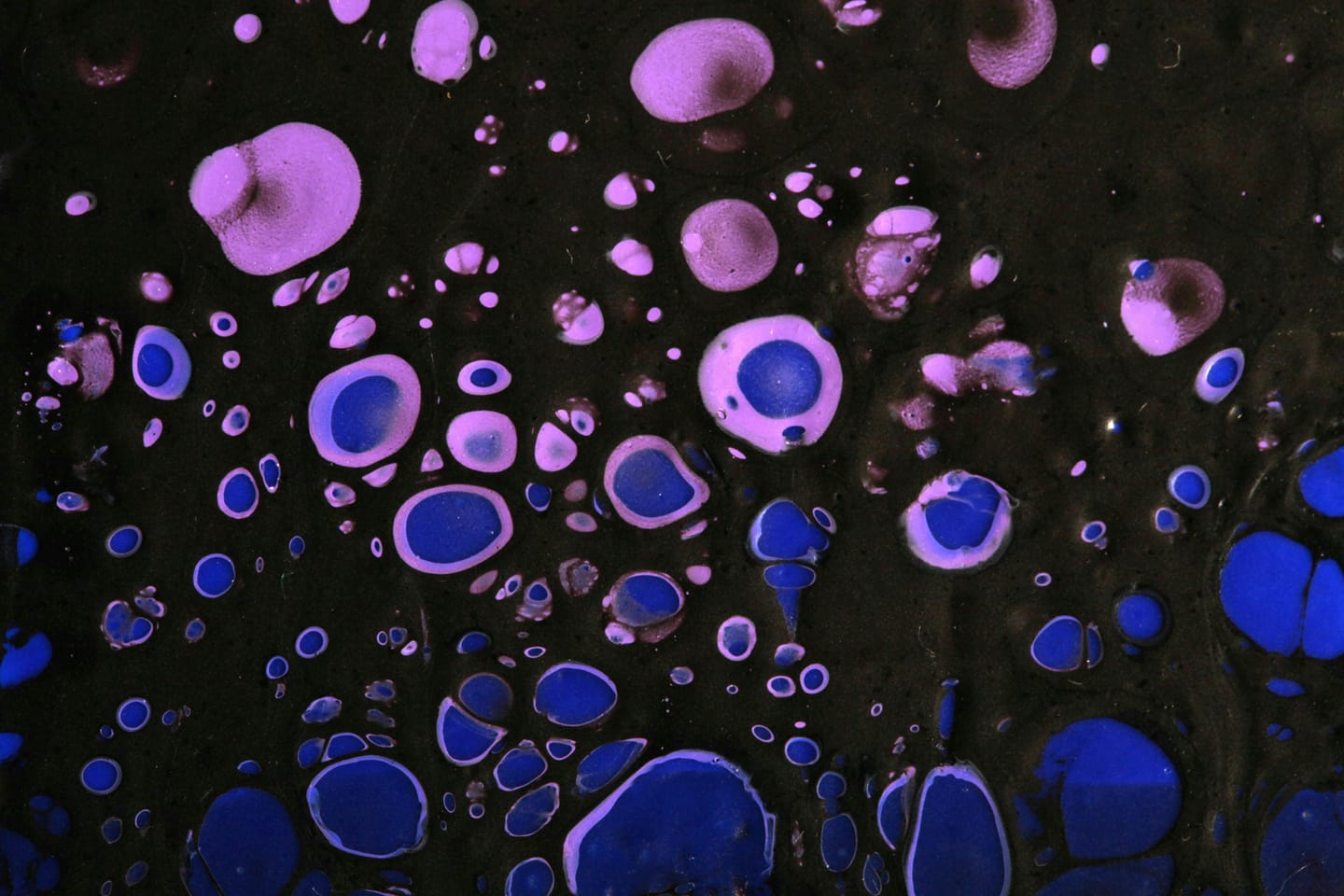
For this experiment, we used microscopy images of Chinese Hamster Ovary (CHO) cells from the BBBC030 dataset — part of the Broad Bioimage Benchmark Collection. This open-access repository provides well-annotated biological image sets designed for benchmarking image analysis methods in bioinformatics and computational biology. The CHO images in BBBC030 present a realistic challenge for segmentation, featuring varying contrast, overlapping cells, and occasional noise or artefacts. These characteristics make the dataset particularly suitable for testing both prompt-based approaches and classical image processing pipelines under realistic conditions.

To evaluate the prompt-based segmentation capabilities of the model, we selected image 12 from the BBBC030 dataset.
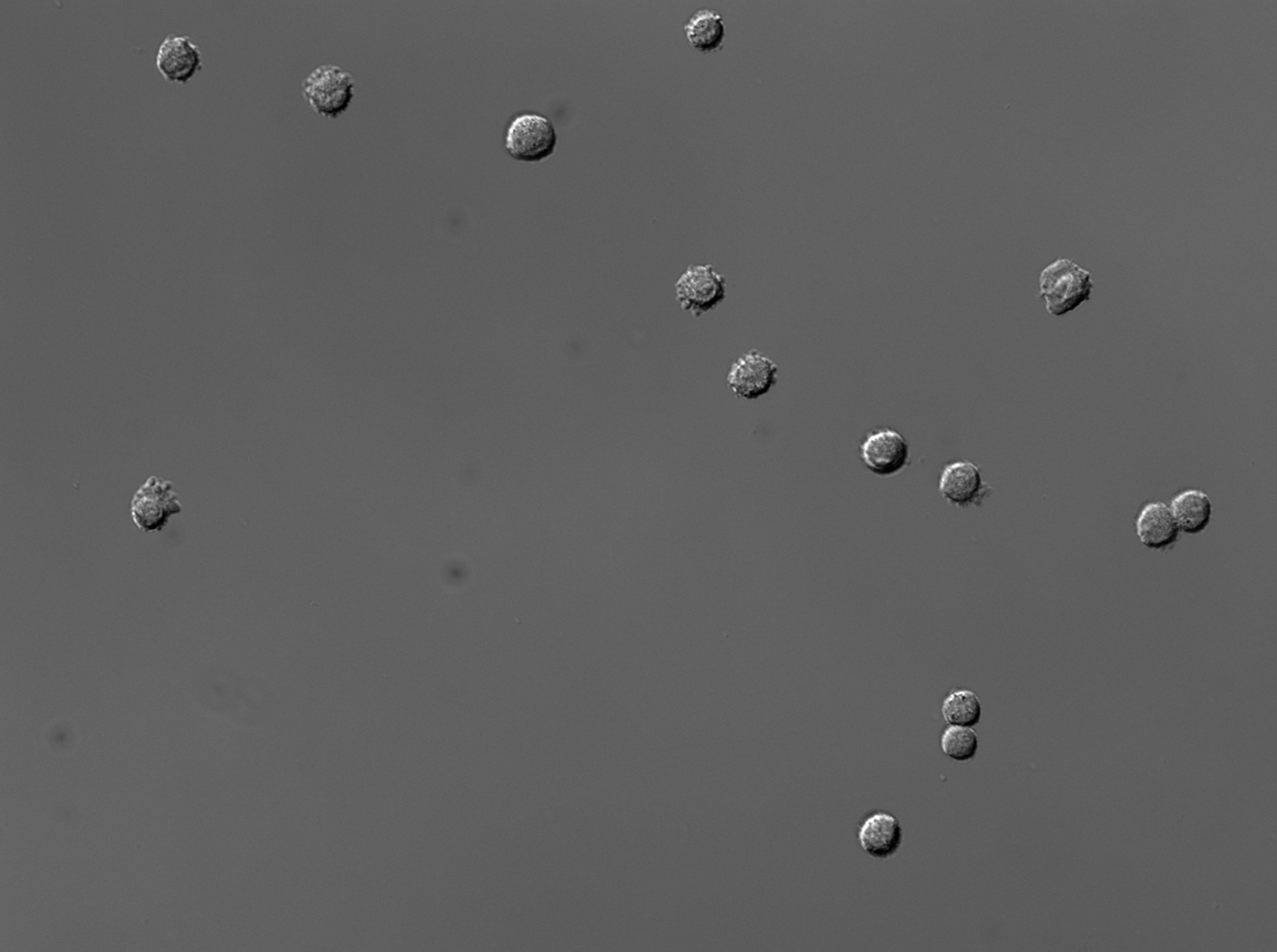
Prompt
This task represents a classical image segmentation problem in bioimage analysis: identifying and isolating the full extent of each visible cell in a microscopy image. The goal was to produce a binary mask in which each complete cell appears as a fully filled white region, and the background remains entirely black. Such masks are crucial for downstream biological analyses, including cell counting, morphology quantification, and phenotypic screening.
Instead of using traditional image processing or supervised machine learning, the segmentation was framed as a natural language prompt — a textual instruction specifying exactly how the mask should look. This allowed us to test whether OpenAI’s o4-mini-high model could translate descriptive instructions into a biologically meaningful output, without any training on biomedical data.
TASK: Generate a filled binary segmentation mask from the microscopy image.
Instructions:
- For each visible cell, generate a filled white mask region that covers the entire area of the cell, from edge to edge.
- The white mask must match the complete shape of each cell — not just its centre, core, or brightest part.
- Each cell must be represented as one solid, filled, connected white blob (pixel value 255).
- The boundary of each cell should be respected, and the entire cell interior should be included.
- Do not include only partial regions. Do not leave out darker parts of the cell.
- The background must remain completely black (pixel value 0).
- Ignore all non-cell objects: dust, shadows, artefacts, specks, and small bright noise-like features.
- Exclude all small white blobs that are significantly smaller than actual cells (e.g., < X pixels), as these are not cells.
- Only retain regions whose size and shape are consistent with real cells.
Output:
- Binary PNG mask.
- Cells = fully filled white areas (255), matching their true size and shape.
- Background = black (0).
- The result must show each cell completely covered, from its outer contour inward — not just highlighted areas, and must exclude small noise specks.
Output
The model followed a multi-step visual reasoning process to solve the segmentation task. It began by analysing the original image and applying Gaussian blur followed by Otsu's thresholding to enhance the contrast between cells and background. Recognising that cells are round and bright, it refined the result with morphological operations such as closing and hole-filling to ensure solid interior masks.

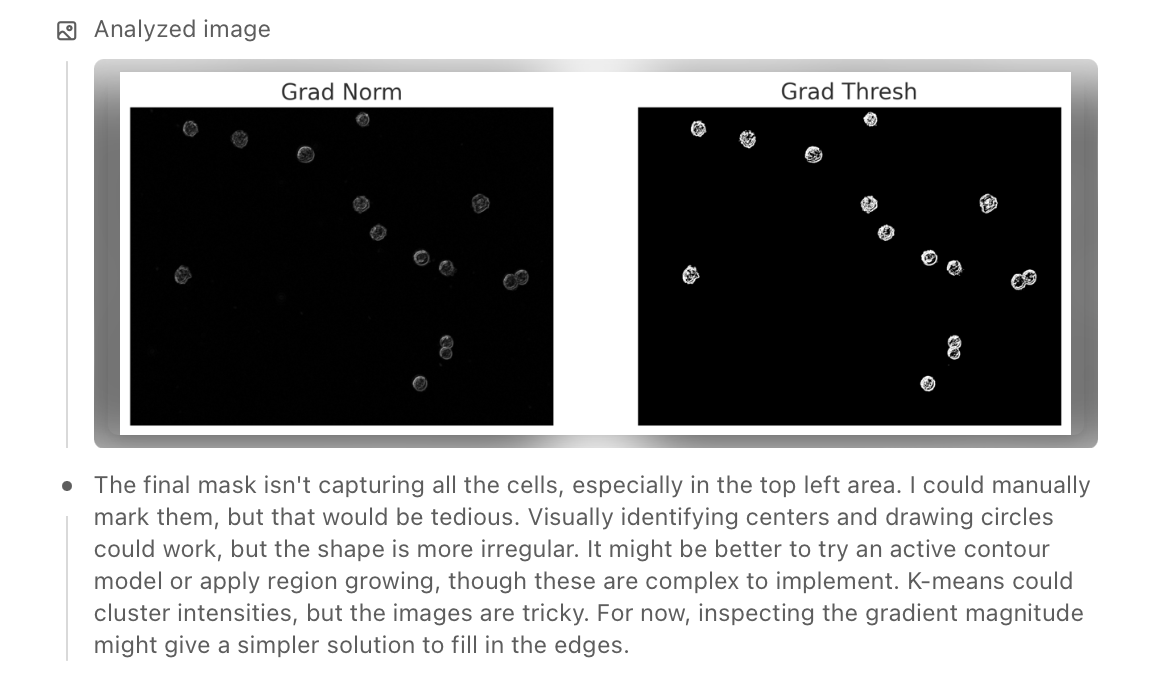
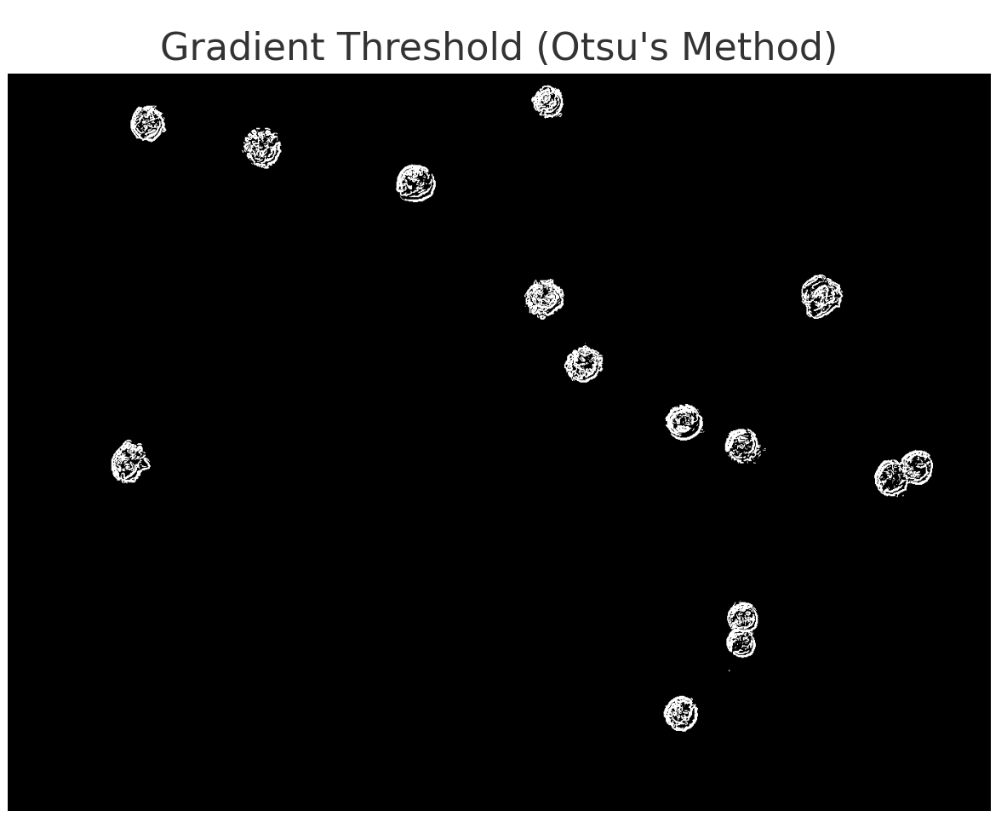
To eliminate noise and non-cell artefacts, the model used connected component analysis, filtering out regions that were either too small or inconsistent in shape. It tested several strategies — including gradient-based filtering, region growing, and distance transforms — before settling on watershed-based segmentation, which helped separate touching cells and improved boundary precision.

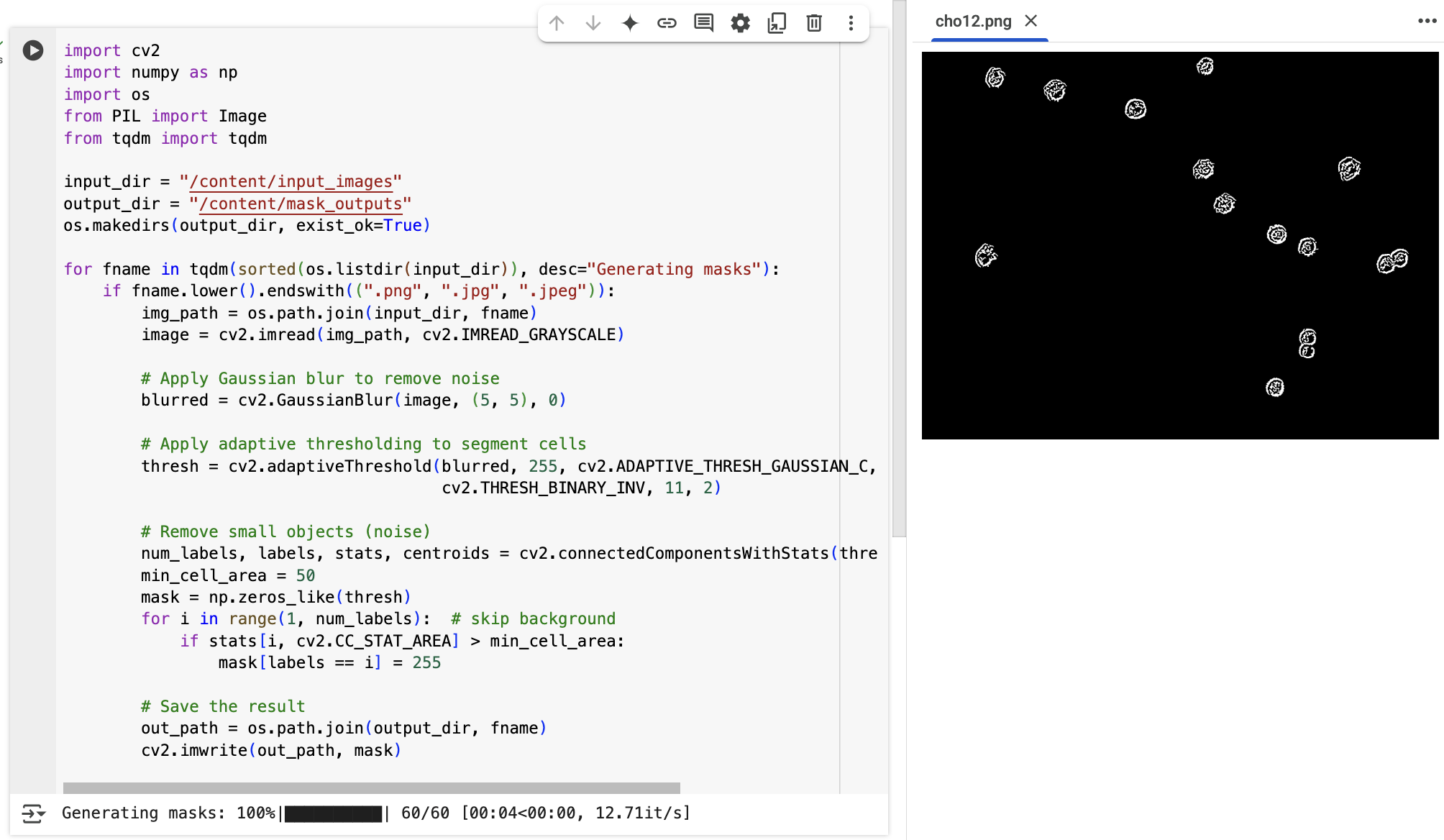
Finally, the model zoomed out, manually verified the detected objects against the original image, and counted the visible cells. It overlaid the mask to check alignment, successfully identifying around 13–15 cells while avoiding most background specks and irrelevant textures.

The resulting binary mask was accurate and clean: the cells were fully enclosed as solid white blobs, the background remained noise-free, and false positives were effectively excluded. This outcome showed that prompt-based reasoning with o4-mini-high could approximate classical image processing pipelines — all without domain-specific training.

In the video below, we overlaid the original microscopy image with the segmentation mask produced by the prompt-based method. The alignment is visually precise — each detected cell matches its real-world position perfectly. This confirms that the resulting binary mask is not only accurate but also reliable for downstream applications. Such masks are commonly used in quantitative cell analysis tasks, including cell counting, tracking, morphological measurements, and training datasets for supervised learning in biomedical imaging.
Script
For users with programming experience, we also applied a classic image processing pipeline to the full dataset of 60 microscopy images using Python and OpenCV. Interestingly, the steps followed in the script mirror the reasoning process demonstrated by the o4-mini-high model — including Gaussian blurring to reduce noise, adaptive thresholding to separate cells from the background, and filtering out small connected components. In other words, the model's prompt-based output reflects a structured, logic-driven segmentation pipeline that can also be reproduced programmatically. This makes it a compelling benchmark for validating prompt-based AI reasoning against traditional code-based methods.
Recommendations
Based on our findings, prompt-based segmentation with OpenAI’s o4-mini-high model offers a lightweight and accessible alternative for generating cell masks in microscopy images. It performs well without requiring domain-specific training or custom code, making it particularly useful for rapid prototyping, educational purposes, or low-resource environments. The results are accurate enough to support downstream tasks such as cell counting, morphology analysis, or dataset preparation for supervised learning. While multiple images (up to ten) can be processed in parallel, the approach still requires prompt-based interaction for each batch, which limits its practicality for fully automated, large-scale pipelines. For high-throughput image segmentation, traditional script-based methods or trained models remain the more scalable and efficient alternative.
Note: Do not use GenAI models for clinical diagnosis or medical decision-making!
The authors used OpenAI o4-mini-high [OpenAI (2025), o4-mini-high (accessed on 2 June 2025), Large language model (LLM), available at: https://openai.com] to generate the output.