Identifying specific clinical findings in unstructured medical texts is a common challenge in healthcare data science. In this post, we benchmark Google’s Gemini 2.5 Flash language model on a zero-shot classification task: detecting the presence or absence of distal fibula fractures in real-world German radiology reports. Without any examples or domain-specific training, Gemini 2.5 Flash correctly classified all 50 reports, demonstrating that large language models can accurately process and label non-English clinical free text in a purely zero-shot setting.
Input file
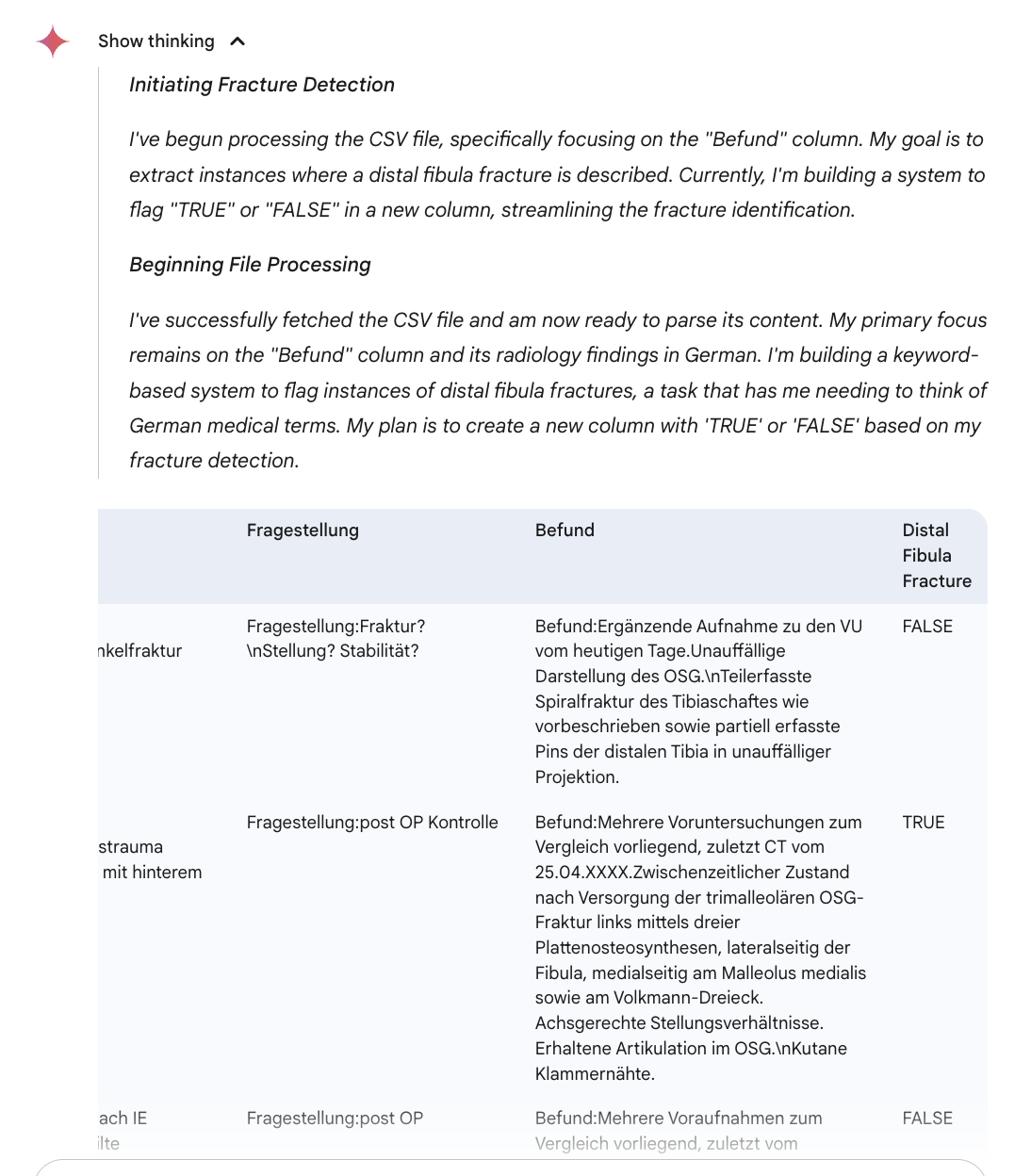
For this experiment, we used a publicly available dataset of German radiology reports from Hannover Medical School, accessible here. From the full collection, we drew a random sample of 50 reports and removed the original binary label column ("Fibula-Fraktur"). The resulting data for each case included four fields: an ID, a composite report text (“Befundtext”), clinical information (“Klin_Angabe”), the clinical question (“Fragestellung”), and the radiologist’s findings (“Befund”). For example, one record from the dataset is structured as follows:

Prompt
The model was instructed to review each radiology report and determine whether the findings described a fracture of the distal fibula, assigning a binary label (“TRUE” or “FALSE”) for each entry. No examples or explanatory notes were provided; the task required the model to interpret the German clinical free text without prior domain adaptation or further guidance. Our aim was to test whether Gemini 2.5 Flash could accurately identify reports describing distal fibula fractures using only the plain text—without any training examples or domain-specific instructions—mirroring the typical challenges encountered in biomedical research when working with unlabelled clinical data.
You are a medical expert in radiology report analysis. Below is a table with several columns, including free-text German radiology findings ("Befund").
For each row, assess whether the "Befund" text describes a distal fibula fracture.
If the report mentions a fracture of the distal fibula, label that row as "TRUE" in the last column.
If there is no mention of a distal fibula fracture, label the row as "FALSE" in the last column.
Return the original table with an additional column (F), filling it only with "TRUE" or "FALSE" for each row. Do not add any explanation or comments.
Output
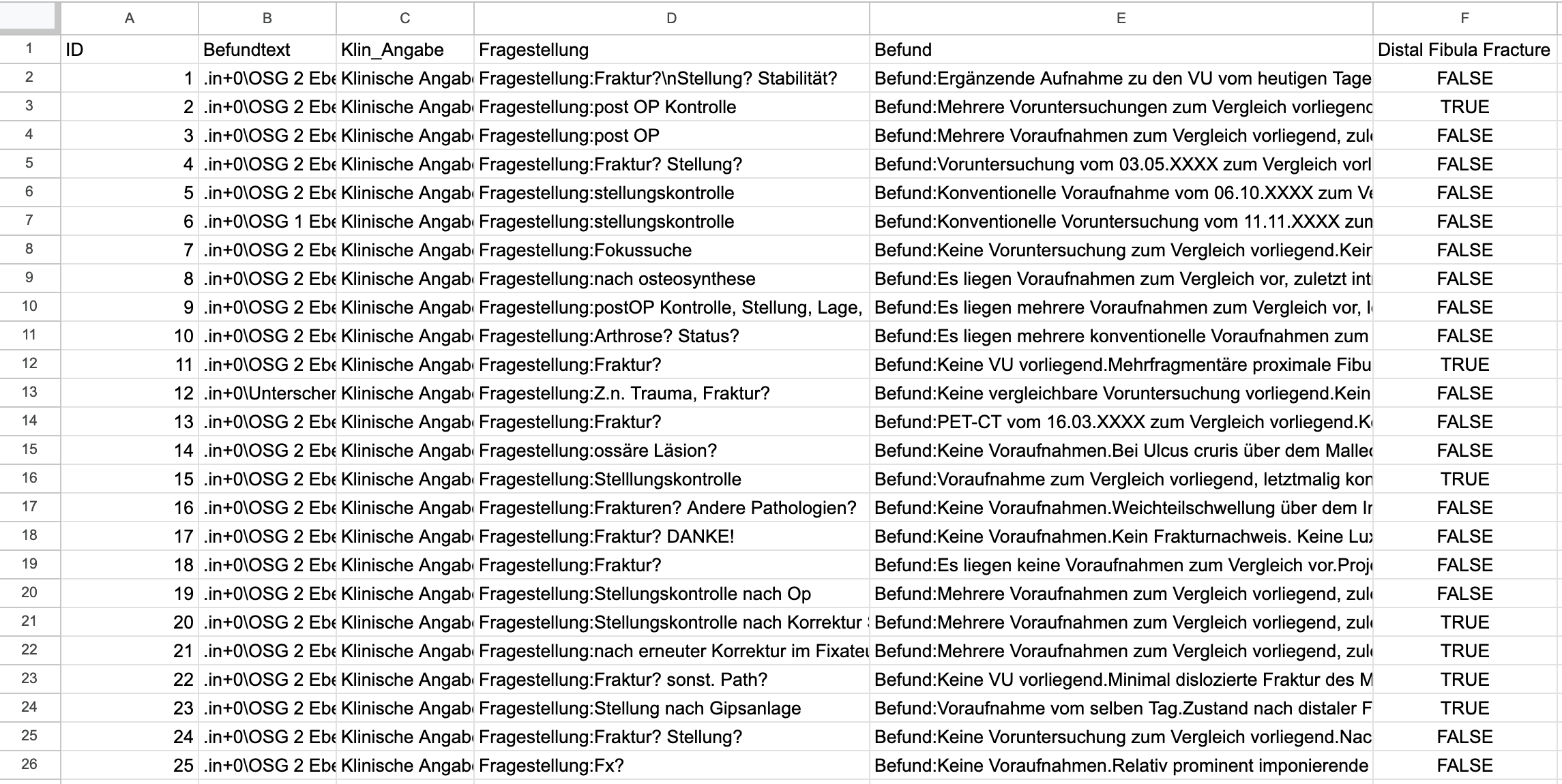
Gemini 2.5 Flash correctly classified all 50 German radiology reports for the presence or absence of a distal fibula fracture, matching the original ground truth labels with complete accuracy. The model’s output was consistent and free from any errors or ambiguous responses, providing a clear binary label (“TRUE” or “FALSE”) for each case. This was achieved without prior exposure to the dataset or language-specific examples. The free-text medical reports included domain-specific terminology and varied phrasing, yet the model handled these consistently.

The model produced results in a format that could be instantly exported to a spreadsheet, allowing for rapid review, sharing, or further analysis within seconds.
Recommendations
Our findings suggest that large language models such as Gemini 2.5 Flash can accurately extract clinically relevant information from unstructured radiology reports, even when working with non-English datasets and without domain-specific training. For biomedical research and clinical audit projects involving free-text clinical records, generative AI models now offer a practical approach for initial data annotation or cohort selection. However, it remains important to validate model outputs against expert review, especially before integrating such results into downstream analyses or clinical workflows.
Note: Do not use GenAI models for clinical diagnosis or medical decision-making!
The authors used Gemini 2.5 Flash [Google DeepMind (2025) Gemini 2.5 Flash (accessed on 2 June 2025), Large language model (LLM), available at: https://deepmind.google/technologies/gemini/] to generate the output.






